

Legumes can interact with rhizobium to form nodules for symbiotic nitrogen fixation (SNF). It is estimated that the fixed N through SNF of legumes accounts for 60-70% of the total biological nitrogen fixation in the world. Therefore, it is of great significance to study the molecular mechanism of legume SNF to improve the SNF capability for the development of green agriculture. SNF efficiency can be affected by legume-rhizobial compatibility and abiotic stresses. However, little is known about their genetic and molecular mechanisms for a long time. Recently, Professor Xuelu Wang's group published two papers on the Nature Plants and Molecular Plant, respectively, to reveal the molecular and evolutionary mechanisms of compatibility for SNF between soybean and rhizobium and the adaptation mechanism of SNF under stress condition. These results provide important theoretical basis and target genes for molecular design breeding of high-efficiency SNF in soybean.
The Professor Xuelu Wang’s group of Henan University and the Professor Youguo Li’s group of Huazhong Agricultural University jointly revealed the genetic, molecular and evolutionary mechanism of the conversion of rhizobia infection mode from intercellular infection to root hair infection during the coevolution of soybean and rhizobia. This study was published on 15th January, 2021 in Nature Plants entitled with "Glycine max NNL1 restrictions symbolic compatibility with bradyrhizobia via root hair infection in Soybean" (doi: 10.1038/s41477-020-00832-7) as research article.
In this paper, first, a soybean core collection contains 496 soybean accessions including 280 landraces and 216 improved cultivars, which has a wide geographical distribution in China, was used for genome-wide association analysis (GWAS). Combining the genotype and phenotype for the nodule number of the core collection after inoculation with Bradyrhizobium diazoefficiens USDA110, a R gene (named GmNNL1, Nodule Number Locus 1) encoding a TIR-NBS-LRR protein, which can restrict the rhizobia infection and reduce the nodule number, was cloned and confirmed by genetic analysis and molecular biology methods.
Further analysis showed that during the natural selection, breeding improvement and coevolution of soybean with rhizobia, the insertion of a GmSINE1 transposon into GmNNL1, led to GmNNL1 protein truncation and inactivation, promoted the recognition of native rhizobia in soil to increase nodule number, and benefited N-fixation activity and biomass accumulation in soybean. The expression of GmNNL1 was specifically induced in root hairs by rhizobia and affected the infection route of rhizobia. The generation of inactivated GmNNL1 haplotype with GmSINE1 insertion plays an important role for the rhizobia infection mode conversion from intercellular infection to root hair infection in soybean. Moreover, through Mass spectrometry analysis, biochemical and genetic analysis, researchers found that the functional full-length GmNNL1 protein can directly interacted with NopP, an effector protein of Bradyrhizobium type III secretion system, to activate ETI, and inhibit the nodulation between soybean and Bradyrhizobium. This study not only revealed the genetic and molecular mechanism of compatibility between soybean and rhizobia for the establishment of a successful nitrogen fixing symbiosis, but also explained conversion event of the major rhizobia infection mode from intercellular infection to efficient root hair infection during the soybean-rhizobia co-evolution. It also provided important theoretical basis and target genes for molecular design breeding of high-efficiency SNF in soybean.
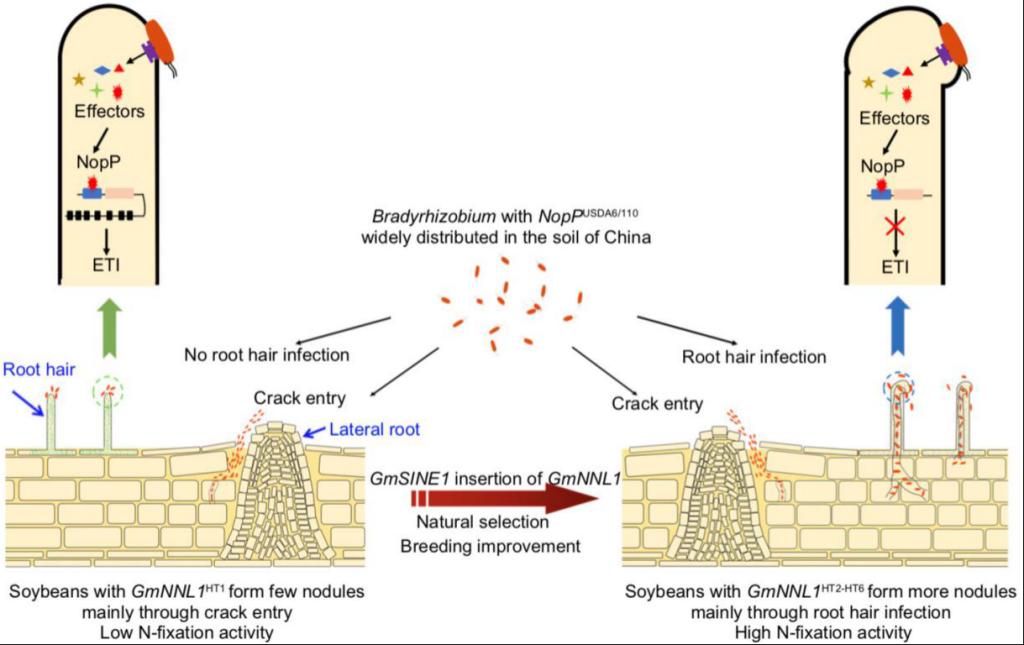
Loss-of-function of GmNNL1 led to the rhizobia infection mode conversion from intercellular infection to efficient root hair infection
On 21th December, 2020, Xuelu Wang’s group published a research article entitled “GSK3-Mediated Stress Signaling Inhibits Legume–Rhizobium Symbiosis by Phosphorylating GmNSP1 in Soybean” on Molecular Plant (doi:10.1016/j.molp.2020.12.015 ). This study revealed that glycogen synthase kinase 3 (GSK3) family genes play an important role in the inhibition of soybean symbiotic signaling and nodule formation under salt stress. GmSK2-8, a GSK3-like kinase induced by salt stress in soybean, interacts with and phosphorylates GmNSP1, a key transcription factor in the symbiotic signaling pathway. The phosphorylation of GmNSP1 inhibits its binding to the promoters of nod factor responsive genes, thereby mediating the salt-inhibited legume–rhizobium symbiosis. This study revealed the molecular mechanism of salt stress regulating nodulation and has important application prospects for improving the efficiency of nitrogen fixation of legumes under environmental stress conditions.
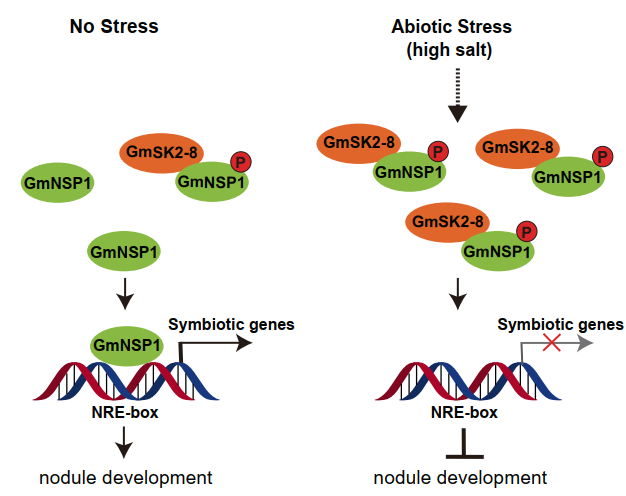
Proposed model of how salt stress inhibits nodulation in soybean
The two studies were supported by the National Key Research and Development Program, the National Natural Science Foundation of China, and the funding of Henan University.
Professor Xuelu Wang’s group has long been engaged in the mechanisms of plant hormone signal transduction, and they regulated plant growth and development under environmental stresses. Recently, Xuelu Wang’s group also made a series of innovative research progresses on signal transduction and plant growth and development. These results included discovering the signal transduction pathways of phytohormone strigolactone and brassinosteroids in regulating shoot branches (Fang et al., Molecular Plant, 2020; Hu et al., Plant Communications, 2020), establishing the spatiotemporal transcriptional regulatory network of brassinosteroids in regulating leaf angle development (Wang et al., iScience, 2020; Wang et al., New Phytologist, 2020), and founding the cellular and molecular mechanisms of root meristem cell differentiation (Chen et al., Journal of Integrative Plant Biology, 2020). Professor Xuelu Wang established the team of "Biological Nitrogen Fixation and Leguminous Biology" at the State Key Laboratory of Crop Adaptation and Improvement in the Henan University. The team focus on the genetics, development, molecular and evolutionary mechanisms of legume-rhizobia symbiosis and important agronomic traits of legume crops. For more information about the "Biological Nitrogen Fixation and Leguminous Biology" team, please visit http://legumelab.com/ .
Contribution: State Key Laboratory of Crop Stress Adaptation and Improvement

 News /
Content
News /
Content


